Molecular Property Prediction and Molecular Design Using a Supervised Grammar Variational Autoencoder
Abstract
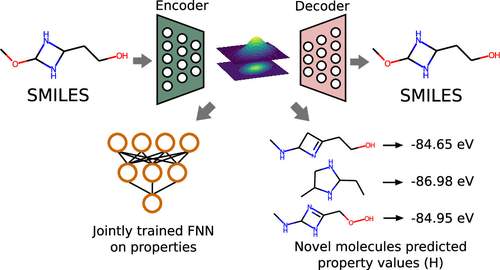
Some of the most common applications of machine learning (ML) algorithms dealing with small molecules usually fall within two distinct domains, namely, the prediction of molecular properties and the design of novel molecules with some desirable property. Here we unite these applications under a single molecular representation and ML algorithm by modifying the grammar variational autoencoder (GVAE) model with the incorporation of property information into its training procedure, thus creating a supervised GVAE (SGVAE). Results indicate that the biased latent space generated by this approach can successfully be used to predict the molecular properties of the input molecules, produce novel and unique molecules with some desired property and also estimate the properties of random sampled molecules. We illustrate these possibilities by sampling novel molecules from the latent space with specific values of the lowest unoccupied molecular orbital (LUMO) energy after training the model using the QM9 data set. Furthermore, the trained model is also used to predict the properties of a hold-out set and the resulting mean absolute error (MAE) shows values close to chemical accuracy for the dipole moment and atomization energies, even outperforming ML models designed to exclusive predict molecular properties using the SMILES as molecular representation. Therefore, these results show that the proposed approach is a viable way to provide generative ML models with molecular property information in a way that the generation of novel molecules is likely to achieve better results, with the benefit that these new molecules can also have their molecular properties accurately predicted.